Presenting at Gain Therapeutics R&D Day, Manolo Bellotto, Ph. D elaborated on the Broad Application of Gain Therapeutics’ Structurally Targeted Allosteric Regulators (STARs). See the presentation video below:
Full Transcript of the Presentation by Manolo Bellotto, Ph. D
We have initially applied SEE-Tx to selective proteins misfolding diseases and namely targeting lysosomal enzyme, addressing the unmet medical needs of rare and neurodegenerative diseases. There are about 50 to 70 different lysosomal storage disorders and only for a portion there are treatments but no cure. This represents a significant addressable market with very high unmet medical needs of a large pediatric population.
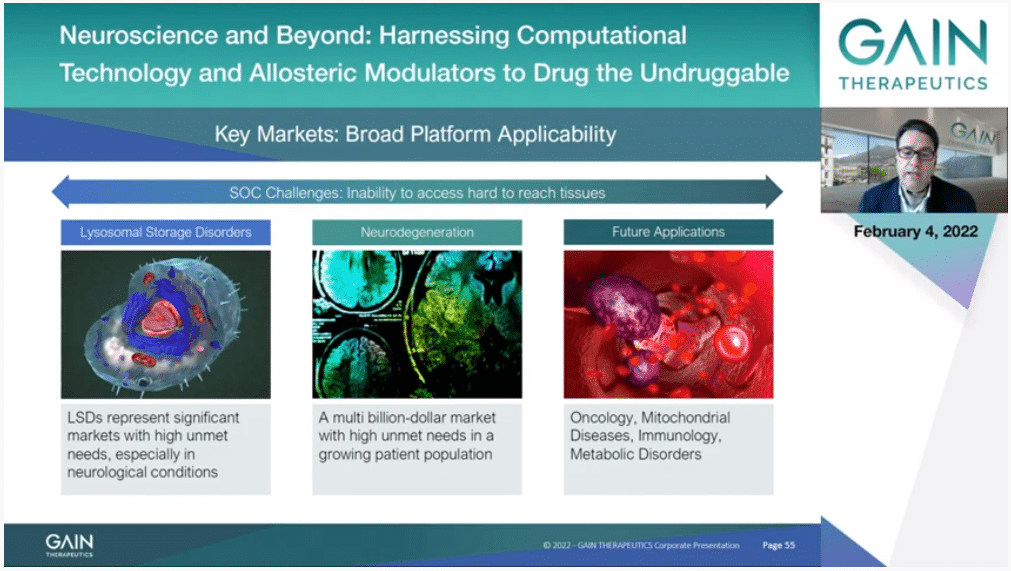
At the same time, a malfunction in lysosomal proteins leads to the molecular onset of some neurodegenerative diseases. Neurodegenerative diseases are large and a growing population where currently there are no disease-modifying therapies on the market, but only symptom relief approaches. Gain is therefore working towards addressing this aspect for some of those disorders.
There are other targets and disorders we are currently working on, including mitochondrial and metabolic disorders. As previously described by Professor Barril and Dr. Broder, there are several targets and molecular aspects of cancer metabolics that deserve new drugs being developed. Thus, we have started entering oncology research as well.
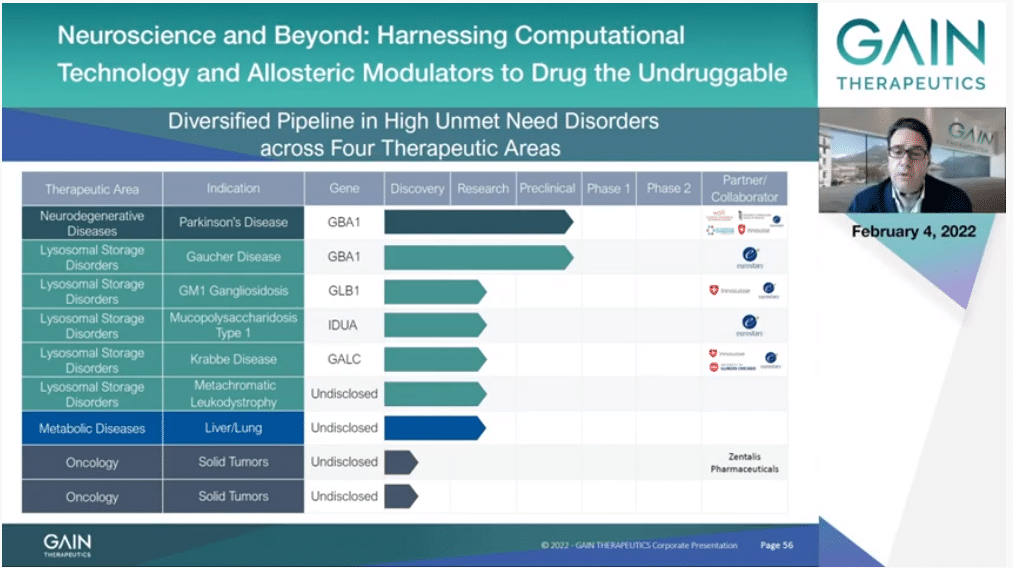
Thanks to SEE-Tx being an agnostic (inaudible) platform technology, we are currently working on four different therapeutic areas addressing the needs of nine different disease indications. Later today I will be presenting our program targeting GBA in Parkinson’s and Gaucher disease, and where we are currently performing new clinical regulatory studies on our candidate drug for Parkinson’s. Additionally to Gaucher, our current programs into lysosomal storage disorder therapeutic field are GM1 gangliosidosis, mucopolysaccharidosis type 1, Krabbe disease and metachromatic leukodystrophy.
In the metabolic therapeutic areas, we have identified a target to address the needs of a rare liver and lung condition.
In oncology, we have identified a relevant oncology target we are currently working on, as mentioned also by Dr. Broder, and are in the process of selecting a new cancer target later this year.
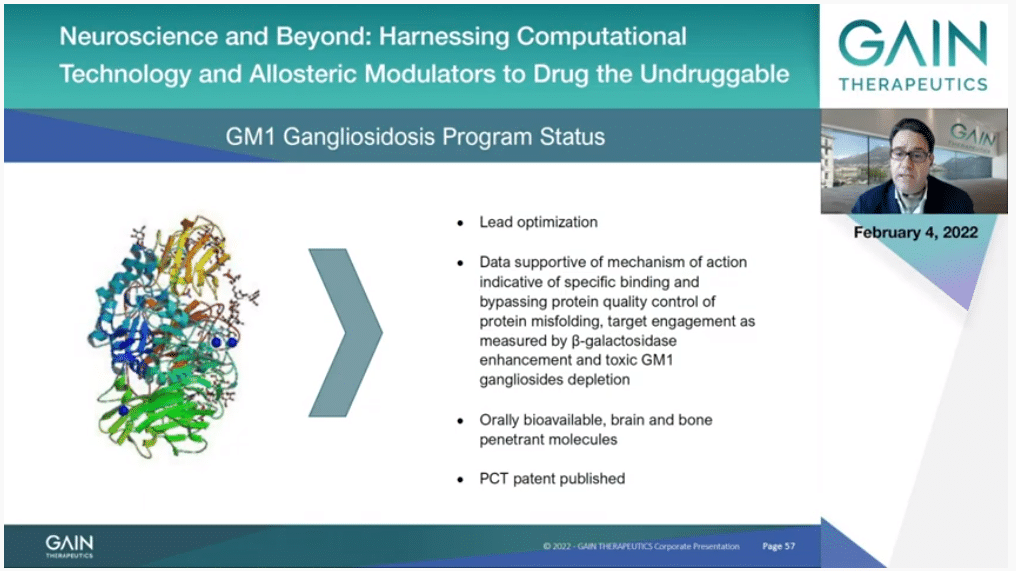
The current status of our GM1 program is that we have entered lead optimization with molecules showing interesting data supportive of beta-galactosidase stabilization, toxic substrate depletion with molecules being orally bioavailable, brain and bone penetrant.
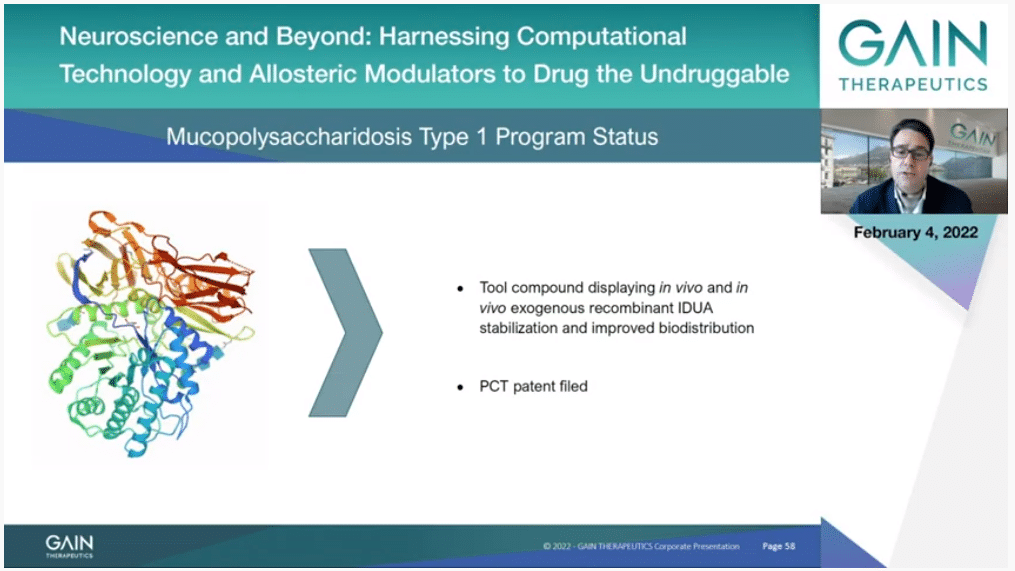
On the next slide, in our mucopolysaccharidosis type 1 program we have identified a tool compound targeting IDUA which is able to stabilize exogenous recombinant IDUA while ameliorating its biodistribution also towards tissue that current medication cannot penetrate.
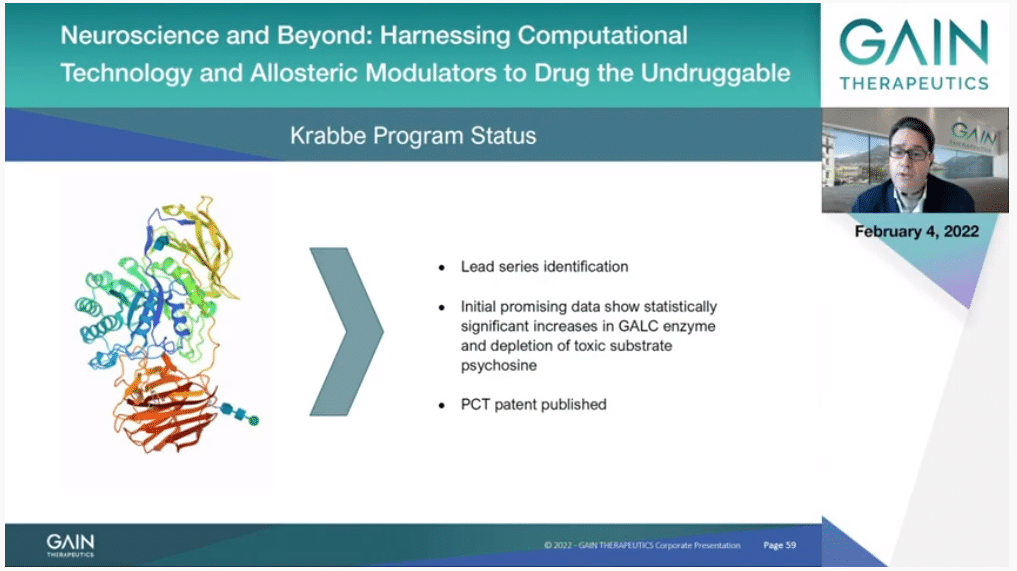
We have started also lead series identification to target GALC for Krabbe disease, and we are happy to report that some early molecules already are showing a positive toxic substrate depletion supportive of the continuation of this program.
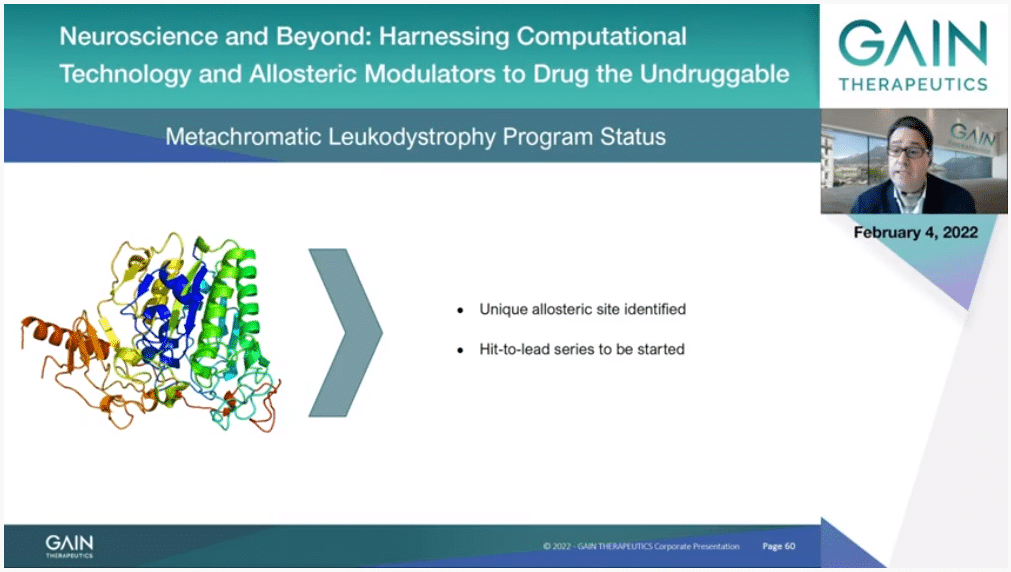
A new program to the Gain pipeline is targeting metachromatic leukodystrophy and where we have already identified a unique allosteric pocket on ARSA.
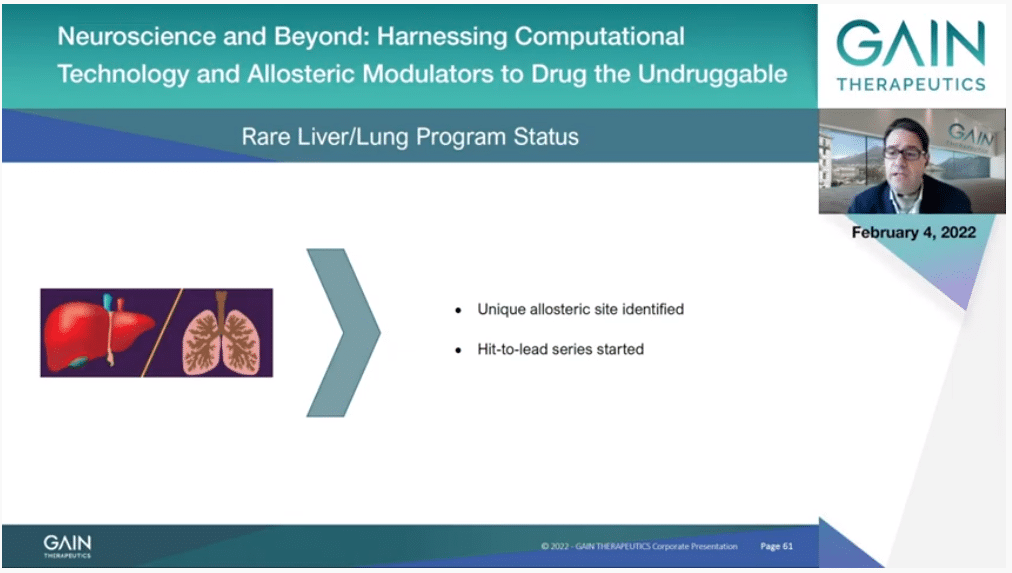
Another new program is targeting a rare liver and lung disorder where we have selected a target on which a unique allosteric site has been identified thanks to SEE-Tx, and where hit-to-lead series has been initiated.
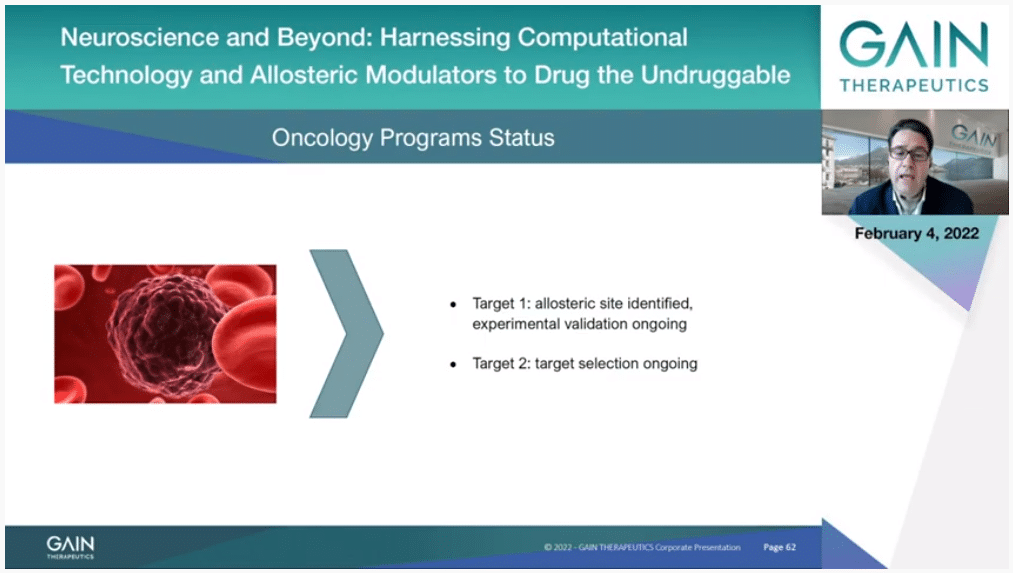
Finally, I would like to introduce to you our new targets in oncology where a unique allosteric site has been identified on a relevant cancer target we are currently not disclosing but where we are performing experimental validation which is ongoing. Additionally, we are in the process of selecting a new target in oncology as well.
As you will appreciate during the course of this R&D Day, SEE-Tx has been generating data validating its approach and this allowed us to broaden its application to other relevant therapeutic areas.
Thank you for your attention.


