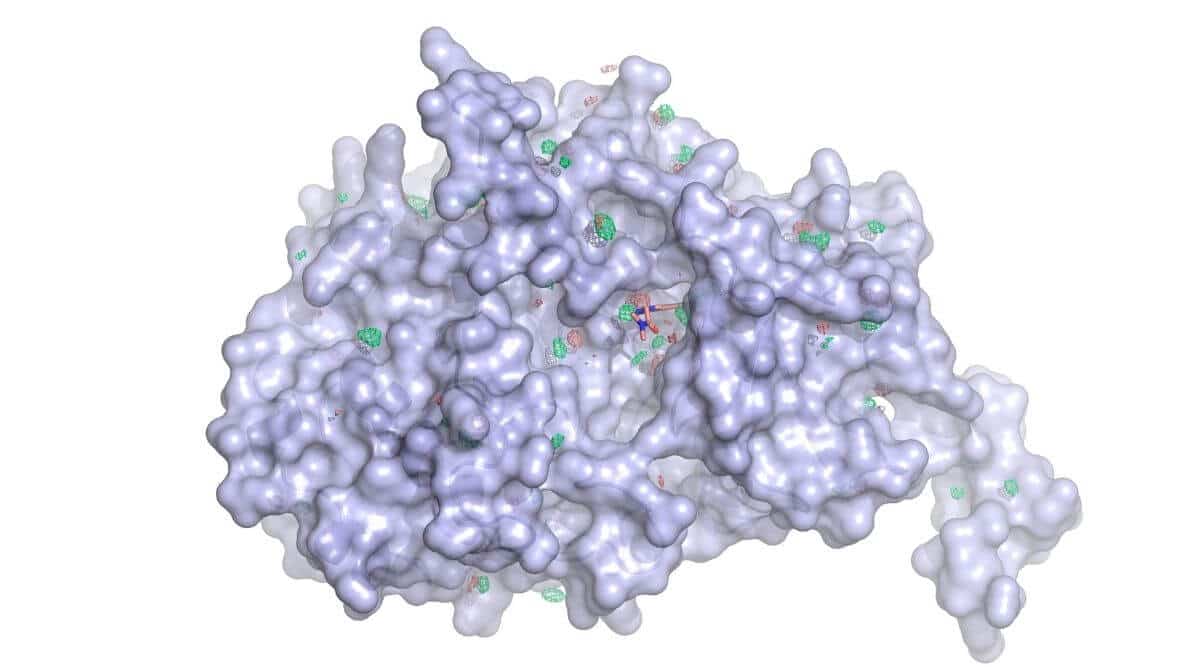
Gain Therapeutics’ physics-based screening platform is able to identify previously unknown binding pockets on well-validated protein targets by identifying cavities on the surface and in crypts that have the potential to be regulatory sites when bound by a small molecule. But what is SEE-TxTM and how does it work?
What is Gain Therapeutics SEE-Tx Platform?
Site-Directed Enzyme Enhancement Therapy (SEE-TxTM) is a physics-based computational platform that uses a proprietary algorithm run on a supercomputer to identify novel drug targets and compounds.
How does Gain Therapeutics SEE-Tx Platform work?
The platform starts with the 3D structure of a well validated protein and runs molecular dynamic simulations. In Molecular Dynamics simulations of mixed organic solvents (MDmix), water and organic solvents compete to interact with the protein surface. Unbiased Molecular Dynamics simulations, followed by density analysis reveal binding hot spots that are clustered into binding sites, irrespective of shape. Deep cavities can result to be undruggable whilst some shallow cavities are druggable. The simulations also take into account protein flexibility and can reveal cryptic sites.
A patented method is used to analyse MDmix results, enabling atomic partitioning of binding free energies and providing quantitative predictions. These values are then used to guide molecular docking in virtual screening and to improve the scores of protein-ligand complexes, resulting in much better success rates.
In the virtual screening process, SEE-Tx filters up to 10 million compounds to select a pool of candidates that may bind to the novel druggable hotspots. Then 100-200 compounds are selected for experimental testing. The final selected small molecules are developed into structurally targeted allosteric regulators (STARs). Gain’s STARs are small molecule therapeutics agents that stabilize misfolded proteins and restore their biological activity.
SEE-Tx TM is able to discover these sites with incredible accuracy and efficiency. Identifying novel allosteric sites on a target enzyme takes 5-6 weeks, compound screening takes, 2-3 weeks, and compound validation can take as little as 2 months.
Read More: AlphaFold Protein Misfolding
Read More: AlphaFold Protein Misfolding Diseases